
Our research focuses on the phytopathogenic bacterium Xanthomonas campestris pv. campestris (Xcc), the causal agent of black rot disease in Brassicaceae. This pathogen infects agriculturally important crops such as cabbage, broccoli, rapeseed and cauliflower, as well as the model plant Arabidopsis thaliana.
Our work focuses on three major processes governing plant–pathogen interactions:
● Plant immune responses to Xanthomonas, especially in hydathodes and the vascular system.
● Genetic basis of Xanthomonas pathogenicity and adaptation to diverse plant tissues and environmental conditions.
● Genetic determinism of host range driven by Xanthomonas' type III effectome.
We employ a wide range of approaches, including molecular genetics, synthetic biology, omics technologies and high-throughput genetic screens to reach these objectives.
Contact: Laurent NOEL
RESEARCH TOPICS
Tissue-specific immune responses of plants to Xanthomonas
Lead researchers: Laurent NOEL et Jean-Marc ROUTABOUL
We explore the molecular, developmental and physiological responses of plants conferring resistance or susceptibility to Xanthomonas infection.
Our work provided an in-depth analysis of hydathode anatomy, physiology, and immunity, highlighting their distinct identity from leaf blades through metabolomic, transcriptomic, and functional genomic approaches. Results showed that hydathodes accumulate auxins, scavenge nutrients, and display unique physiological functions. Additionally, work on cauliflower hydathodes revealed that Xcc type III effectors strongly suppress immune responses in this tissue.
(A) Leaf tooth containing hydathodes and leaf blade samples were macro-dissected from leaves of 9- to 10-week-old plants and subjected to RNA sequencing. (B) Functional categorisation of the 1086 differentially expressed genes with a functional annotation (461/625 induced/repressed in hydathodes). DEGs were mainly annotated using the MapMan analysis classification available from the BAR site . The colour scale indicates the mean log2 Fold Change (FC) of the genes belonging to the given functional category that were globally either more expressed in hydathodes (in red) or in the leaf blade (in blue). (C,D) Fresh cryo-prepared samples observed by cryo-scanning electron microscopy. (E) Observation in Nomarsky of a clarified hydathode sample. Note the numerous xylem vessels irrigating the hydathode.
We explore the response of Arabidopsis conferring resistance or susceptibility to Xanthomonas infection by exploiting forward genetic screens performed with the HEM mutant collection.
(a)Picture of a 104 Hem lines plants out of the 900. (b) Violin plot representation of the distribution of the number of mutations per line
We focus on the immunity established in plant hydathodes and vasculature which impact on pathogen entry and spread, respectively. In particular, we study the role of water availability in the establishment of resistance and susceptibility to Xanthomonas. This axis is supported by two ANR grants (VIP and HIRAQUIM)
Xanthomonas adaptations to plant tissues and temperature
Lead researcher: Alice BOULANGER
Pathogen adaptation to environmental changes is key to virulence. Transcriptomic analyses have provided insights into stress and nutritional status of Xanthomonas at early infection stages and uncovered diverse HrpG-dependent virulence strategies across Xanthomonas strains.
Schematic representation of main transcriptional responses of Xcc happening during the early stage of hydathode colonization: Genes corresponding to blue and red objects are repressed and induced between 4 and 72 hr post inoculation, respectively. Genes corresponding to grey objects are not differentially expressed. T3SS: type III secretion system; T3E: type III effector; T2SS: type II secretion system; LPS: lipopolysaccharide; PCWDE: plant cell wall-degrading enzymes; T4P: type IV pilus. Figure drafted using biorender.
We conduct high-throughput RB-TnSeq screens to identify Xcc genes involved in colonization of plant tissues and adaptation to various environments with a particular focus on hypothetical proteins (HPs) and uncharacterized transcriptional regulators (TRs).
Experimental workflow
Type III-dependent virulence mechanisms and genetic determinism of host range
Lead researchers: Corinne AUDRAN et Carlos ZARATE CHAVES
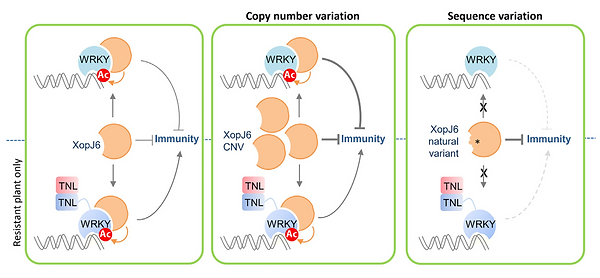
Our team investigates the individual and collective roles of Xanthomonas type III effectors (T3Es). We identified XopJ6, a homolog of Ralstonia pseudosolanacearum PopP2, as a key factor triggering disease resistance in cauliflower and Arabidopsis. Our study revealed how XopJ6 can either trigger or evade plant immunity and how increased copy number of xopJ6 promotes virulence.
In parallel, we engaged into the deconstruction of the entire Xcc effectome yielding an effectorless nonpathogenic strain). Interestingly, its full pathogenicity can be restored on Arabidopsis by a synthetic effectome composed of only six effectors. The team currently implements a synthetic biology pipeline combined with high-throughput fitness measurements to assess mini-effectome functionality on host and nonhost plants.
TALEs (Transcription Activator-Like Effectors) are Xanthomonas-specific T3E proteins that modulate plant gene expression to facilitate infection. Their crucial role in Xanthomonas pathogenesis has been demonstrated using CRISPR interference technology, which allowed precise repression of tal gene expression in different bacterial strains.
In the context of Xcc, the specific contribution of TALEs is now being investigated both in natural host plants and in nonhost species with the aim of identifying their direct plant targets and clarifying their dual role as either virulence or avirulence factors.
By understanding the mechanisms of Xanthomonas virulence and host range, we aim at predicting how these pathogens may emerge in novel environments and adapt to new host plants. This work could lay the groundwork for the development of predictive models that can inform pathogen outbreaks and help mitigate the impact of bacterial diseases in agriculture.
Collaborations
-
Adam Bogdanove, Cornell University, NY
-
Jian-Min Zhou, Beijing, China
-
Jennifer Lewis, UC Berkeley, CA.
-
Anne Genissel, INRA Versailles, France
-
Richard Berthomé, Laurent Deslandes and Fabrice Roux, LIPM Toulouse, France
-
Boris Szurek & Ralf Koebnik, IRD Montpellier, France
-
Matthieu Barret, Nicolas Chen and Marie-Agnès Jacques, INRA Angers, France
-
Lionel Gagnevin and Olivier Pruvost, CIRAD Reunion, France
Ongoing funding
-
ANR VIP (2024-2029) ANR-24-CE20-5527-01 Immunité vasculaire des plantes/plant vascular immunity. L. Noël (Coordinator, LIPME Toulouse), L. Navarro (IBENS Paris), N. Peeters (LIPME Toulouse), Marie-Laure Martin Magnette (IPS2 Saclay). 818k€ (360k€ for LIPME).
-
ANR HIRAQUIM (2023-2028) ANR-23-CE20-0022-02 Rôle régulateur des protéines HIR localisées dans les nanodomaines membranaires dans l'immunité dépendante des aquaporines chez les plantes/Regulatory role of the membrane nanodomain‐localized proteins HIR in the aquaporin‐dependent plant immunity. Enric Zelazny (Coodinator, IPSIM Montpellier), L. Noël (LIPME Toulouse). 547k€ (214k€ for LIPME).
-
INRAE PE Department Funding: XccOnFire (2025-2026), A. Boulanger, 30k €
-
Research networks INRAE SPE Department: FNX = French Network on Xanthomonads. MA Jacques, R. Koebnik, O. Pruvost and L. Noël, coordinators, 5 k € / year.




PUBLICATIONS
2026
Chesneau, G., Noel, A., Bréard, D., Boulanger, A., Briand, M., Bonneau, S., Brin, C., Saux, M. F.-L., Liu, Y., Hendrickson, A., Nielsen, T., Sarniguet, A., Guilet, D., Arkin, A., Lui, L., & Barret, M. (2026). Lactuchelins represent lipopeptide siderophores produced by Pseudomonas lactucae that inhibit Xanthomonas campestris. The ISME Journal, wrag003. https://doi.org/10.1093/ismejo/wrag003
2025
Martínez Rivas, F. J., Wozny, D., Xue, Z., Gilbault, E., Sapir, T., Rouille, M., Ricou, A., Medina, J., Noël, L. D., Lauber, E., Voxeur, A., Mazier, M., Loudet, O., Clément, G., & Jiménez-Gómez, J. M. (2025). Parallel evolution of salinity tolerance in Arabidopsis thaliana accessions from Cape Verde Islands. Science Advances, 11(28), eadq8210. https://doi.org/10.1126/sciadv.adq8210
Routaboul, J.-M., Corso, M., & Lepiniec, L. (2025). Flavonoid characterization : It takes more than Arabidopsis seed colours! Planta, 262(5), 112. https://doi.org/10.1007/s00425-025-04828-5
2024
Routaboul, J.-M., Bellenot, C., Olympio, A., Clément, G., Citerne, S., Remblière, C., Charvin, M., Franke, L., Chiarenza, S., Vasselon, D., Jardinaud, M.-F., Carrère, S., Nussaume, L., Laufs, P., Leonhardt, N., Navarro, L., Schattat, M., & Noël, L. D. (2024). Arabidopsis hydathodes are sites of auxin accumulation and nutrient scavenging. The Plant Journal: For Cell and Molecular Biology. https://doi.org/10.1111/tpj.17014
Quiroz Monnens, T., Roux, B., Cunnac, S., Charbit, E., Carrère, S., Lauber, E., Jardinaud, M.-F., Darrasse, A., Arlat, M., Szurek, B., Pruvost, O., Jacques, M.-A., Gagnevin, L., Koebnik, R., Noël, L. D., & Boulanger, A. (2024). Comparative transcriptomics reveals a highly polymorphic Xanthomonas HrpG virulence regulon. BMC Genomics, 25(1), 777.
https://doi.org/10.1186/s12864-024-10684-6
Carrère, S., Routaboul, J.-M., Savourat, P., Bellenot, C., López, H., Sahoo, A., Quiroz Monnens, T., Ricou, A., Camilleri, C., Declerck, N., Laufs, P., Mercier, R., & Noël, L. D. (s. d.). A fully sequenced collection of homozygous EMS mutants for forward and reverse genetic screens in Arabidopsis thaliana. The Plant Journal, 2024 Jul 29. https://doi.org/10.1111/tpj.16954
Koebnik, R., Cesbron, S., Chen, N. W. G., Fischer-Le Saux, M., Hutin, M., Jacques, M.-A., Noël, L. D., Perez-Quintero, A., Portier, P., Pruvost, O., Rieux, A., & Szurek, B. (2024). Celebrating the 20th Anniversary of the First Xanthomonas Genome Sequences – How Genomics Revolutionized Taxonomy, Provided Insight into the Emergence of Pathogenic Bacteria, Enabled New Fundamental Discoveries and Helped Developing Novel Control Measures – A Perspective from the French Network on Xanthomonads. Zenodo. https://doi.org/10.5281/zenodo.10683038
Lauber, E., González-Fuente, M., Escouboué, M., Vicédo, C., Luneau, J. S., Pouzet, C., Jauneau, A., Gris, C., Zhang, Z.-M., Pichereaux, C., Carrère, S., Deslandes, L., & Noël, L. D. (2024). Bacterial host adaptation through sequence and structural variations of a single type III effector gene. iScience, 27(3), Article 3. https://doi.org/10.1016/j.isci.2024.109224
Monnens, T. Q., & Boulanger, A. (2024). A large scale bacterial attraction assay : A new quantitative bacterial migration assay suitable for genetic screens. PLOS ONE, 19(6), e0305037. https://doi.org/10.1371/journal.pone.0305037
2023
Marzorati, F., Rossi, R., Bernardo, L., Mauri, P., Silvestre, D. D., Lauber, E., Noël, L. D., Murgia, I., & Morandini, P. (2023). Arabidopsis thaliana Early Foliar Proteome Response to Root Exposure to the Rhizobacterium Pseudomonas simiae WCS417. Molecular Plant-Microbe Interactions: MPMI, 36(11), 737‑748. https://doi.org/10.1094/MPMI-05-23-0071-R
Talbi, N., Fokkens, L., Audran, C., Petit-Houdenot, Y., Pouzet, C., Blaise, F., Gay, E. J., Rouxel, T., Balesdent, M.-H., Rep, M., & Fudal, I. (2023). The neighbouring genes AvrLm10A and AvrLm10B are part of a large multigene family of cooperating effector genes conserved in Dothideomycetes and Sordariomycetes. Molecular Plant Pathology. https://doi.org/10.1111/mpp.13338
Zárate-Chaves, C. A., Audran, C., Medina Culma, C. A., Escalon, A., Javegny, S., Gagnevin, L., Thomas, E., Pimparé, L.-L., López, C. E., Jacobs, J. M., Noël, L. D., Koebnik, R., Bernal, A. J., & Szurek, B. (2023). CRISPRi in Xanthomonas demonstrates functional convergence of transcription activator-like effectors in two divergent pathogens. The New Phytologist, 238(4), 1593‑1604. https://doi.org/10.1111/nph.18808
2022
You, Y., Koczyk, G., Nuc, M., Morbitzer, R., Holmes, D. R., von Roepenack-Lahaye, E., Hou, S., Giudicatti, A., Gris, C., Manavella, P. A., Noël, L. D., Krajewski, P., & Lahaye, T. (2022). The eINTACT system dissects bacterial exploitation of plant osmosignalling to enhance virulence. Nature Plants, 1‑14. https://doi.org/10.1038/s41477-022-01302-y
Luneau, J. S., Baudin, M., Quiroz Monnens, T., Carrère, S., Bouchez, O., Jardinaud, M.-F., Gris, C., François, J., Ray, J., Torralba, B., Arlat, M., Lewis, J. D., Lauber, E., Deutschbauer, A. M., Noël, L. D., & Boulanger, A. (2022). Genome-wide identification of fitness determinants in the Xanthomonas campestris bacterial pathogen during early stages of plant infection. The New Phytologist. https://doi.org/10.1111/nph.18313
Bellenot, Caroline, Jean-Marc Routaboul, Patrick Laufs, et Laurent D. Noël. « Hydathodes ». Current Biology 32, no 14 (25 juillet 2022): R763‑64. https://doi.org/10.1016/j.cub.2022.06.014.
Bellenot, C., Carrère, S., Gris, C., Noël, L. D., & Arlat, M. (2022). Genome Sequences of 17 Strains from Eight Races of Xanthomonas campestris pv. Campestris. Microbiology Resource Announcements, 11(7), e0027922. https://doi.org/10.1128/mra.00279-22
Luneau, J. S., Noël, L. D., Lauber, E., & Boulanger, A. (2022). A β-glucuronidase (GUS) Based Bacterial Competition Assay to Assess Fine Differencesin Fitness during Plant Infection. Bio-protocol, 12(13), e3776. https://doi.org/10.21769/BioProtoc.3776
Dubrow, Zoe, Sara Carpenter, Morgan E. Carter, Ayress Grinage, Carine Gris, Emmanuelle Lauber, Jules Butchacas, et al. « Cruciferous Weed Isolates of Xanthomonas Campestris Yield Insight into Pathovar Genomic Relationships and Genetic Determinants of Host- and Tissue-Specificity ». Molecular Plant-Microbe Interactions: MPMI, 10 mai 2022. https://doi.org/10.1094/MPMI-01-22-0024-R.
Luneau, Julien S., Aude Cerutti, Brice Roux, Sébastien Carrère, Marie-Françoise Jardinaud, Antoine Gaillac, Carine Gris, et al. « Xanthomonas Transcriptome inside Cauliflower Hydathodes Reveals Bacterial Virulence Strategies and Physiological Adaptations at Early Infection Stages ». Molecular Plant Pathology 23, no 2 (février 2022): 159‑74. https://doi.org/10.1111/mpp.13117.
2021
Zhu, Xiaoyang, Julie Mazard, Eugénie Robe, Sarah Pignoly, Marielle Aguilar, Hélène San Clemente, Emmanuelle Lauber, Richard Berthomé, et Jean-Philippe Galaud. « The Same against Many: AtCML8, a Ca2+ Sensor Acting as a Positive Regulator of Defense Responses against Several Plant Pathogens ». International Journal of Molecular Sciences 22, no 19 (28 septembre 2021): 10469. https://doi.org/10.3390/ijms221910469.
2020
Jauneau, Alain, Aude Cerutti, Marie-Christine Auriac, et Laurent D. Noël. « Anatomy of Leaf Apical Hydathodes in Four Monocotyledon Plants of Economic and Academic Relevance ». PloS One 15, no 9 (2020): e0232566. https://doi.org/10.1371/journal.pone.0232566.
González-Fuente, Manuel, Sébastien Carrère, Dario Monachello, Benjamin G. Marsella, Anne-Claire Cazalé, Claudine Zischek, Raka M. Mitra, et al. « EffectorK, a Comprehensive Resource to Mine for Ralstonia, Xanthomonas, and Other Published Effector Interactors in the Arabidopsis Proteome ». Molecular Plant Pathology 21, no 10 (octobre 2020): 1257‑70. https://doi.org/10.1111/mpp.12965.
Gluck-Thaler, Emile, Aude Cerutti, Alvaro L. Perez-Quintero, Jules Butchacas, Verónica Roman-Reyna, Vishnu Narayanan Madhavan, Deepak Shantharaj, et al. « Repeated Gain and Loss of a Single Gene Modulates the Evolution of Vascular Plant Pathogen Lifestyles ». Science Advances 6, no 46 (novembre 2020): eabc4516. https://doi.org/10.1126/sciadv.abc4516.
Arroyo-Velez, Noe, Manuel González-Fuente, Nemo Peeters, Emmanuelle Lauber, et Laurent D. Noël. « From Effectors to Effectomes: Are Functional Studies of Individual Effectors Enough to Decipher Plant Pathogen Infectious Strategies? » PLoS Pathogens 16, no 12 (décembre 2020): e1009059. https://doi.org/10.1371/journal.ppat.1009059.
2019
Cerutti, Aude, Alain Jauneau, Patrick Laufs, Nathalie Leonhardt, Martin H. Schattat, Richard Berthomé, Jean-Marc Routaboul, et Laurent D. Noël. « Mangroves in the Leaves: Anatomy, Physiology, and Immunity of Epithemal Hydathodes ». Annual Review of Phytopathology 57 (25 août 2019): 91‑116. https://doi.org/10.1146/annurev-phyto-082718-100228.
2018
Denancé, Nicolas, Boris Szurek, Erin L. Doyle, Emmanuelle Lauber, Lisa Fontaine-Bodin, Sébastien Carrère, Endrick Guy, et al. « Two Ancestral Genes Shaped the Xanthomonas Campestris TAL Effector Gene Repertoire ». The New Phytologist 219, no 1 (juillet 2018): 391‑407. https://doi.org/10.1111/nph.15148.
2017
Cerutti, Aude, Marie-Christine Auriac, Laurent D. Noël, et Alain Jauneau. « Histochemical Preparations to Depict the Structure of Cauliflower Leaf Hydathodes ». Bio-Protocol 7, no 20 (20 octobre 2017): e2452. https://doi.org/10.21769/BioProtoc.2452.
Cerutti, Aude, et Alain Jauneau. « Capturing Z-Stacked Confocal Images of Living Bacteria Entering Hydathode Pores of Cauliflower ». Bio-Protocol 7, no 20 (20 octobre 2017): e2451. https://doi.org/10.21769/BioProtoc.2451.
Cerutti, Aude, Alain Jauneau, Marie-Christine Auriac, Emmanuelle Lauber, Yves Martinez, Serge Chiarenza, Nathalie Leonhardt, Richard Berthomé, et Laurent D. Noël. « Immunity at Cauliflower Hydathodes Controls Systemic Infection by Xanthomonas Campestris Pv Campestris ». Plant Physiology 174, no 2 (juin 2017): 700‑716. https://doi.org/10.1104/pp.16.01852.
2016
Boulanger, Alice, et Laurent D. Noël. « Xanthomonas Whole Genome Sequencing: Phylogenetics, Host Specificity and Beyond ». Frontiers in Microbiology 7 (2016): 1100. https://doi.org/10.3389/fmicb.2016.01100.
Denancé, Nicolas, Thomas Lahaye, et Laurent D. Noël. « Editorial: Genomics and Effectomics of the Crop Killer Xanthomonas ». Frontiers in Plant Science 7 (2016): 71. https://doi.org/10.3389/fpls.2016.00071.
Jacques, Marie-Agnès, Matthieu Arlat, Alice Boulanger, Tristan Boureau, Sébastien Carrère, Sophie Cesbron, Nicolas W. G. Chen, et al. « Using Ecology, Physiology, and Genomics to Understand Host Specificity in Xanthomonas ». Annual Review of Phytopathology 54 (4 août 2016): 163‑87. https://doi.org/10.1146/annurev-phyto-080615-100147.
Robène, I., S. Bolot, O. Pruvost, M. Arlat, L. D. Noël, S. Carrère, M.-A. Jacques, R. Koebnik, et L. Gagnevin. « High-Quality Draft Genome Sequences of Two Xanthomonas Pathotype Strains Infecting Aroid Plants ». Genome Announcements 4, no 5 (1 septembre 2016): e00902-16. https://doi.org/10.1128/genomeA.00902-16.
2015
Bolot, Stéphanie, Aude Cerutti, Sébastien Carrère, Matthieu Arlat, Marion Fischer-Le Saux, Perrine Portier, Stéphane Poussier, Marie-Agnes Jacques, et Laurent D. Noël. « Genome Sequences of the Race 1 and Race 4 Xanthomonas Campestris Pv. Campestris Strains CFBP 1869 and CFBP 5817 ». Genome Announcements 3, no 5 (17 septembre 2015): e01023-15. https://doi.org/10.1128/genomeA.01023-15.
Pesce, Céline, Stéphanie Bolot, Edwige Berthelot, Claude Bragard, Sébastien Cunnac, Marion Fischer-Le Saux, Perrine Portier, et al. « Draft Genome Sequence of Xanthomonas Translucens Pv. Graminis Pathotype Strain CFBP 2053 ». Genome Announcements 3, no 5 (8 octobre 2015): e01174-15. https://doi.org/10.1128/genomeA.01174-15.
Pesce, Céline, Stéphanie Bolot, Sébastien Cunnac, Perrine Portier, Marion Fischer-Le Saux, Marie-Agnès Jacques, Lionel Gagnevin, et al. « High-Quality Draft Genome Sequence of the Xanthomonas Translucens Pv. Cerealis Pathotype Strain CFBP 2541 ». Genome Announcements 3, no 1 (12 février 2015): e01574-14. https://doi.org/10.1128/genomeA.01574-14.
Roux, Brice, Stéphanie Bolot, Endrick Guy, Nicolas Denancé, Martine Lautier, Marie-Françoise Jardinaud, Marion Fischer-Le Saux, et al. « Genomics and Transcriptomics of Xanthomonas Campestris Species Challenge the Concept of Core Type III Effectome ». BMC Genomics 16 (18 novembre 2015): 975. https://doi.org/10.1186/s12864-015-2190-0.
Wang, Guoxun, Brice Roux, Feng Feng, Endrick Guy, Lin Li, Nannan Li, Xiaojuan Zhang, et al. « The Decoy Substrate of a Pathogen Effector and a Pseudokinase Specify Pathogen-Induced Modified-Self Recognition and Immunity in Plants ». Cell Host & Microbe 18, no 3 (9 septembre 2015): 285‑95. https://doi.org/10.1016/j.chom.2015.08.004.












